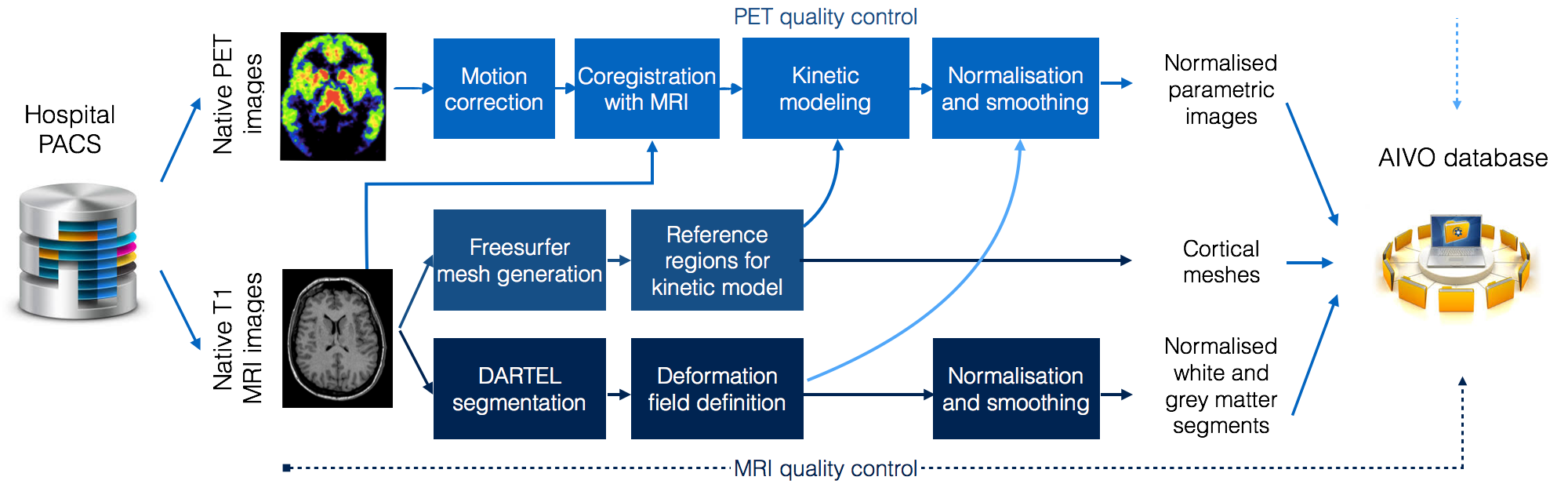
Integrated brain image processing pipeline at Turku PET Centre
Traditional preprocessing PET data requires substantial manpower and is subject to human error, consequently this approach is not suitable for large-scale, standardized analyses. To resolve this issue, the AIVO data are processed using the MAGIA preprocessing pipeline developed at the Human Emotion Systems Laboratory, Turku PET Centre (principal developer MSc Tech Tomi Karjalainen). The MAGIA implements automatic image retrieval from PACS combined with functions from FreeSurfer and SPM image preprocessing pipelines coupled with in-house PET modelling and metadata annotation code. Quality control is performed with in-house code and MRIQC toolbox. The compilation of MAGIA code is site-independent and can be installed anywhere from the git page.
Kinetic modelling
The MAGIA system currently has implementations for the follwing PET models which can all be used depending on the type of PET data that are available
- Simplified reference tissue model (Gunn et al., 1997)
- Patlak Graphical plot model with reference tissue and blood input (Patlak et al, 1983)
- Fractional Uptake Rate (Camici et al., 1986)
- SUV ratio (Hamberg et al., 1994)
- Logan plot (Logan et al., 1990)
The Turku PET Centre has an excellent overview of the different modelling approaches (curated by Vesa Oikonen) which you may consult for further information regarding the models.
Input
Because MAGIA is aimed at processing both controlled scientific as well as “wild type” clinical neuroimaging data, the system is flexible with respect to the input data. Optimally, both dynamic PET scans, input data, as well as isotropic T1 with MPRAGE or equivalent sequence should be available to allow automated reference tissue definition and normalization. If MRI is not availbale, reference tissue definition and normalization are based on atlases. If istatic PET images are used, modeling can still be performed with FUR or SUV ratio.
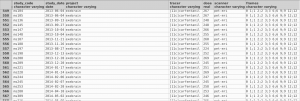
MAGIA processing starts by entering the key subject and scan information (file locations, names, frames, injected doses, desired model etc) into the data processing frame. When all is set, MAGIA will start churning the files and amend the data processing frame with the output and quality control metrics. This allows convenient management of data from multiples studies as well as large-scale data integration.
Output
The MAGIA system will automatically output and store
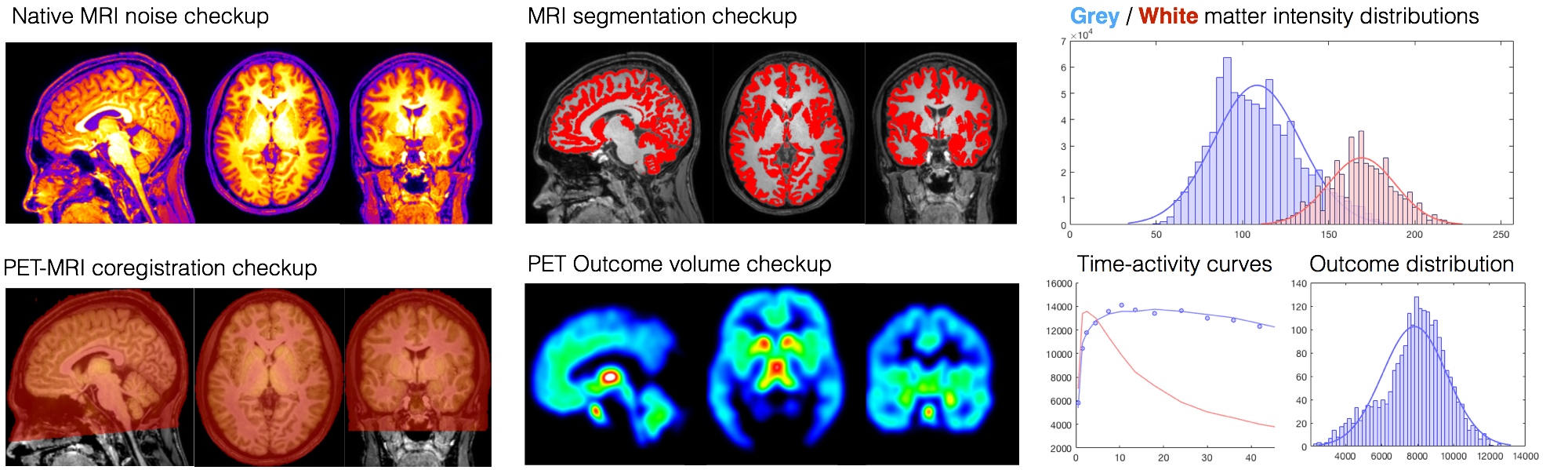
- Modelled PET images, with and without normalization and smoothing (depending on the implemented kinetic model)
- Time-activity curves and input functions
- FreeSurfer surface meshes if T1 image was available
- Grey and white matter segments based on the Dartel pipeline the if T1 image was available
- Visual and numerical quality control metrics and processing specs
Database interaction
The real advantage of this system is that int interacts seamlessly with PostgrSQL database (although MAGIA works as a standalone too), thus all the metadata for the processed images as well as the outptuts can be stored centrally. This allows efficient storage, quality assurance and large-scale analyses of images with the stored metadata.
Download
To download and install the MAGIA pipeline, please click here for our GIT page, which also contains instructions for setting up the pipeline and running various preprocessing streams.